In this post I’ll walk through pulling COVID-19 data, cleaning it, and then visualizing it.
The datasets for COVID-19 are publicly available and updated daily which makes them a great candidate for analysis. Since the charts are posted daily in the news, this project offers an easy way to double-check one’s work! Lastly, since the pandemic is still in full force in a handful of countries, new trends are emerging every day from these datasets so there is always something interesting to explore.
My main goals with this project include:
- Connect to a data source and automatically pull the latest data whenever the script is run
- Clean the data and convert it from aggregate numbers to new daily totals
- Filter the data to see the countries that have the highest case counts
- Chart the top 20 countries with the most number of new daily cases
- Repeat the same data cleaning steps with the State Data and chart them to see the trends
To start, I’ll import the libraries we need for the analysis and the source data. I didn’t find one source for all the data so I pulled in the country data from John’s Hopkins and the state data from the New York Times. Using the requests library, I can grab the latest uploaded CSV and save it to my working directory.
import datetime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import calendar
import urllib.request
# counrty data from johns hopkins
url = 'https://data.humdata.org/hxlproxy/api/data-preview.csv?url=https%3A%2F%2Fraw.githubusercontent.com%2FCSSEGISandData%2FCOVID-19%2Fmaster%2Fcsse_covid_19_data%2Fcsse_covid_19_time_series%2Ftime_series_covid19_confirmed_global.csv&filename=time_series_covid19_confirmed_global.csv'
urllib.request.urlretrieve(url, '/working_directory/time_series_covid19_confirmed_global.csv')
#state data from new york times
url_states = 'https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-states.csv'
urllib.request.urlretrieve(url_states, '/working_directory/covid_datasets/us-states.csv')
countries_df = pd.read_csv('covid_datasets/time_series_covid19_confirmed_global.csv')
states_df = pd.read_csv('covid_datasets/us-states.csv')Let’s preview the country data:

A few observations:
- The data is pivoted. We can tell this because it has the dates as column headers and new entries will be posted as a new column. I’ll use the
pd.meltfunction later on to unpivot the data - The data is cumulative, meaning this is showing the total number of coronavirus cases as of that date. I’ll use the
df.diff()function later on to find the new cases per day - Lastly, the Province/State Column is mostly empty and will cause problems later on unless we remove it
I’m going to run df.describe to get an idea of how many countries are in the dataset.

Since there are 266 countries, that may cause a lot of noise when charting. So, I plan to sort the data by the largest 7-day change and grab the top 20 countries.
countries_df['7 day change'] = countries_df.iloc[:,-1] / (countries_df.iloc[:,len(countries_df.columns) - 7]) -1
I’m also going to add a ranking column.
countries_df['Rank'] = countries_df['7 day change'].rank(ascending = False)
The next thing I want to do is to turn the cumulative data into new daily cases. I’ll be taking advantage of the pandas.diff() function to accomplish this.
#use the diff to turn the cumulative data into new daily cases. diff can only be run on the number columns so I can isolate those by using iloc diff_df = countries_df.iloc[:,4:-2].diff(axis=1) #Isolating just the countrie columns, the 7 day change column, and the rank column countries_only_df = countries_df.iloc[:, np.r_[:4,-2,-1]] #Merging the dataframes back together merge1 = pd.concat([countries_only_df,diff_df], axis = 1, join = 'inner') #Creating a new dataframe that is sorted by yesterdays number in daily cases country_new_daily_cases = merge1.sort_values(by=merge1.columns[-1], ascending = False)
Next, I’ll isolate the top 20 countries.
top_20_countries = country_new_daily_cases[:20]
Now, I can un-pivot the data using the pandas.melt() function.
df_new = pd.melt(frame=top_20_countries, id_vars=['Country/Region','Lat','Long','7 day change','Rank'], value_vars=date_column, value_name="Number of Cases", var_name="Date")
Some data clean up with the date and creating a modified country name column.
#Converting the date to a datetime
df_new['Date'] = pd.to_datetime(df_new['Date'], format='%m/%d/%y')
#Creating a new column that combines the country name with the % Change to use in the title of the subplot
df_new['Country (7 day change)']= df_new["Country/Region"] + ' (' + (round((df_new["7 day change"]*100),1)).map(str) + '%)' + " (Rank "+df_new["Rank"].astype(int).map(str) + ')'Let’s plot!
sns.set()
sns.set_style("whitegrid")
sns.set_context("poster", font_scale = 1)
f, ax = plt.subplots(figsize=(20,15))
sns.lineplot(data = df_new, x='Date', y='Number of Cases', hue = 'Country/Region')
plt.legend(ncol = 5,loc = (0,1.1))
xmax= df_new['Date'].max()
filt = df_new[df_new.Date == df_new.Date.max()]
plt.tight_layout()
plt.savefig('new_daily_cases.png')
plt.show()
A few observations:
- The data varies a lot day to day
- It’s hard to tell what’s going on with so much variability
To solve this, I am going to find the 7-day rolling average by using the .rolling() function in Pandas. I am going to use the pivoted dataframe from earlier right after I calculated the new daily cases.
rolling_avg_df = diff_df.rolling(window=7, axis = 1).mean() merge2 = pd.concat([countries_only_df,rolling_avg_df], axis = 1, join = 'inner') country_new_daily_cases_rolling_avg = merge2.sort_values(by=merge2.columns[-1], ascending = False) top_20_countries_roll = country_new_daily_cases_rolling_avg[:20] countries_rolling_average = pd.melt(frame=top_20_countries_roll, id_vars=['Country/Region','Lat','Long'], value_vars=date_column, value_name="Number of Cases", var_name="Date") countries_rolling_average['Date'] = pd.to_datetime(countries_rolling_average['Date'], format='%m/%d/%y')
Now we can chart it.
# get a closer look at the lower countries
sns.set()
sns.set_style("whitegrid")
sns.set_context("poster", font_scale = 1)
f, ax = plt.subplots(figsize=(20,13))
sns.lineplot(data = countries_rolling_average, x='Date', y='Number of Cases', hue = 'Country/Region')
plt.legend(ncol = 4,loc = (0,1.1))
plt.title('Coronavirus Cases by Country - 7 Day Moving Average')
xmax= df_new['Date'].max()
#return only the first 4 countries labels becasue the rest just overlap
filt = countries_rolling_average[df_new.Date == df_new.Date.max()][:4]
for index, row in filt.iterrows():
ax.annotate(row['Country/Region'], xy=(row['Date'], row['Number of Cases']),
xycoords='data')
f.tight_layout()
plt.savefig('covid_cases_by_country.png')
plt.show()

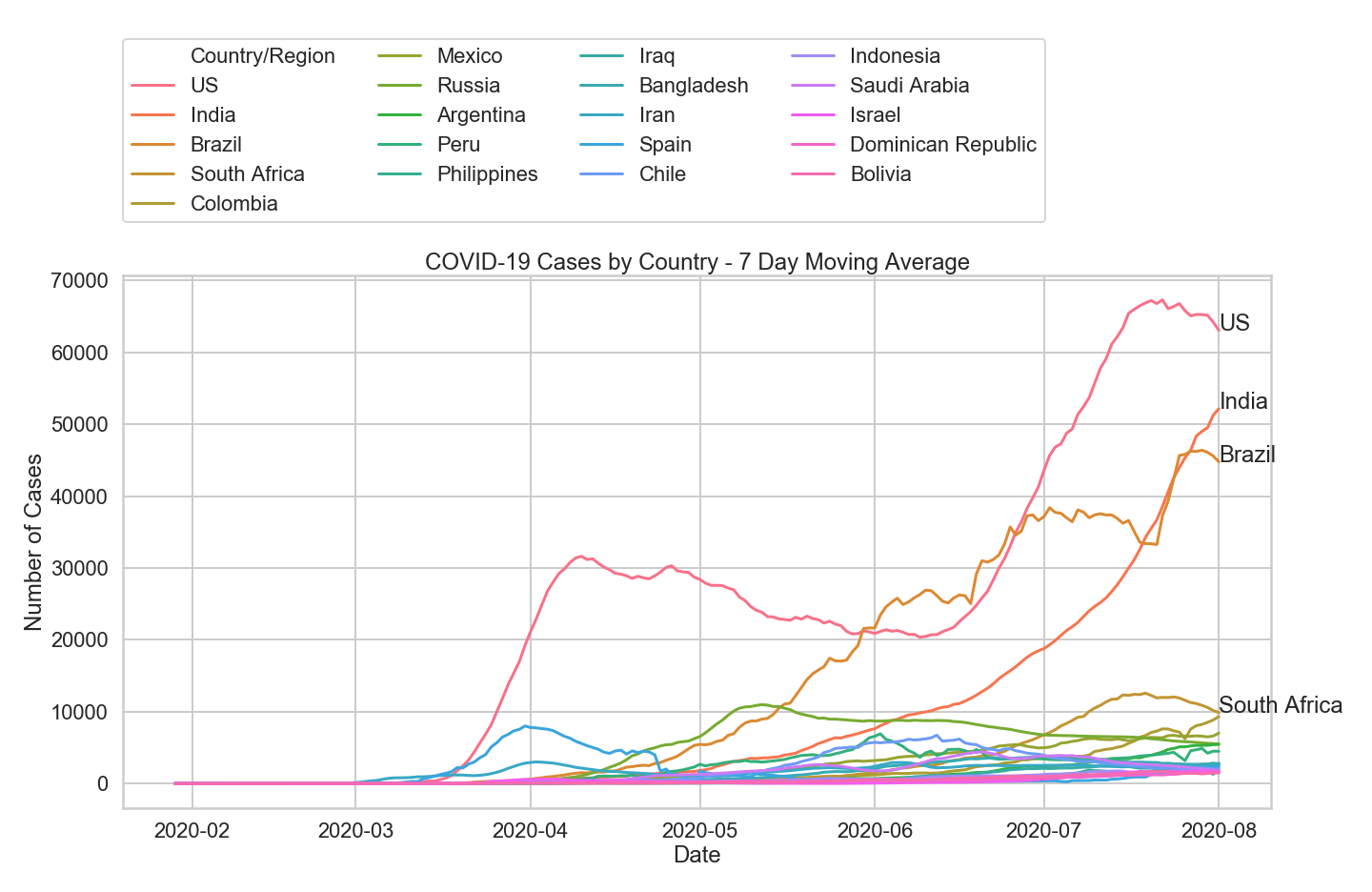
Much easier to read. A few observations:
- The US leads the world in new daily cases
- Brazil was the leader in new cases in June
- India recently overtook Brazil as the country with the second highest case count. Their trajectory also follows a standard exponential curve
- Most countries have less than 10,000 new cases per day
Because there are so many countries between 0 to 10,000 new daily cases, it is not easy to tell what their trajectory looks like. Are they coming down? Going up? To isolate each individual country, I can iterate through the top 20 to make an individual subplot for each.
fig, axes = plt.subplots(10,2, figsize=(100,100))
for (year, group), ax in zip(df_new.groupby('Country (7 day change)'), axes.flatten()):
group.plot(x='Date', y='Number of Cases', kind='line', ax=ax, title=year)
fig.tight_layout()
plt.savefig('cases_by_country_individual.png')
Now we can see that in Argentina, for example, cases look to be increasing exponentially.
State Data
Now on to the state data.
states_df = pd.read_csv('target_directory/us-states.csv')
states_dfTaking a quick preview of the data

The data is already tidy which is great. However, I have a feeling it’s also cumulative data. Running a quick plot using my home state confirms my suspicion. Minnesota doesn’t have over 50,000 new daily cases, the line is too smooth to represent new daily cases, and a quick Google search for new daily cases also shows me what I would expect to see if the data was not cumulative.

To calculate the new daily cases, I will pivot the table and then take the difference from the previous days total.
states_pivoted = pd.pivot_table(states_df, values='cases', index='state',columns='date', aggfunc=np.sum, fill_value=0) states_diff_df = states_pivoted.diff(axis=1) states_rolling_df = states_diff_df.rolling(window=7, axis = 1).mean() states_rolling_df = states_rolling_df.reset_index() new_states_rolling_df = pd.melt(frame = states_rolling_df.reset_index(), id_vars='state', value_vars=states_rolling_df.columns[1:], value_name="Number of Cases", var_name="Date") #Convert the date from an object to a datetime new_states_rolling_df['Date'] = pd.to_datetime(new_states_rolling_df['Date'], format='%Y-%m-%d')
Next, I want to see the highest number of cases a state experienced. This will be used for my data labels when I chart this.
state_worst_day = new_states_rolling_df.loc[new_states_rolling_df.reset_index().groupby(['state'])['Number of Cases'].idxmax()].sort_values(by="Number of Cases", ascending=False) state_worst_day
Let’s plot!
sns.set()
sns.set_style("whitegrid")
sns.set_context("poster", font_scale = 1)
f, ax = plt.subplots(figsize=(20,9))
sns.lineplot(data = new_states_rolling_df, x='Date', y='Number of Cases', hue = 'state')
#plt.legend(ncol = 5,loc = (0,1.1))
plt.title('Coronavirus Cases by US State - 7 Day Moving Average')
date_filter = new_states_rolling_df['Date'] == new_states_rolling_df['Date'].max()
state_labels = new_states_rolling_df.loc[date_filter].sort_values(by="Number of Cases", ascending = False)[:4]
#Top 4 States
for index, row in state_labels.iterrows():
ax.annotate(row['state'], xy=(row['Date'], row['Number of Cases']),
xycoords='data')
#Add Data Labels for specific states with select states with a spike in cases
select_states = ["New York", "New Jersey", "Arizona", "Illinois"]
for i in select_states:
ax.annotate(i, xy=(state_worst_day.loc[state_worst_day['state'] == i]['Date'],state_worst_day.loc[state_worst_day['state'] == i]['Number of Cases']))
#ax.annotate("New Jersey", xy=(state_worst_day.loc[state_worst_day['state'] == 'New Jersey']['Date'],state_worst_day.loc[state_worst_day['state'] == 'New Jersey']['Number of Cases']))
ax.get_legend().remove()
#f.tight_layout()
plt.savefig('covid_cases_by_state.png')
plt.show()

Like I mentioned earlier, there are a ton of ways to display the data as well as a lot of insights into trends. Hopefully, this allows you to get started playing with the dataset. Check out my full Jupyter Notebook on my GitHub